Gene Expression In Eukaryotes Vs Prokaryotes
Muz Play
Mar 18, 2025 · 6 min read
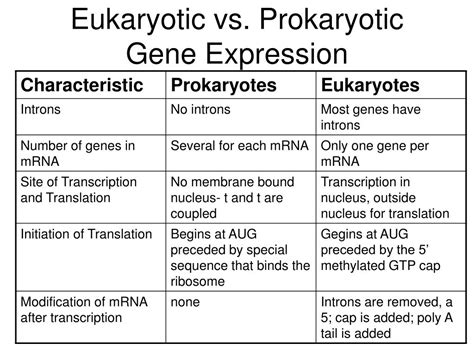
Table of Contents
Gene Expression: A Tale of Two Domains – Eukaryotes vs. Prokaryotes
Gene expression, the intricate process of converting genetic information encoded in DNA into functional proteins, differs significantly between eukaryotes and prokaryotes. While both share the fundamental steps of transcription (DNA to RNA) and translation (RNA to protein), the complexities introduced by the eukaryotic cellular architecture lead to a vastly more regulated and nuanced process. This article delves into the key distinctions in gene expression between these two domains of life, exploring the mechanisms, regulatory elements, and evolutionary implications.
Transcriptional Differences: The Nucleus Plays a Crucial Role
One of the most striking differences lies in the location of transcription. In prokaryotes, like bacteria and archaea, transcription and translation occur simultaneously in the cytoplasm. The absence of a nucleus means that ribosomes can directly bind to mRNA molecules even as they are being transcribed, leading to rapid protein synthesis. This coupled transcription-translation is crucial for the swift response of prokaryotes to environmental changes.
Eukaryotes, on the other hand, possess a membrane-bound nucleus that physically separates transcription from translation. Transcription occurs within the nucleus, producing a pre-mRNA molecule. This pre-mRNA undergoes extensive processing before it can be exported to the cytoplasm for translation. This spatial separation allows for multiple layers of gene regulation that are simply not possible in prokaryotes.
Pre-mRNA Processing: A Multi-Step Refinement
The processing of eukaryotic pre-mRNA involves several key steps:
-
5' Capping: A 7-methylguanosine cap is added to the 5' end of the pre-mRNA. This cap protects the mRNA from degradation and is crucial for its recognition by the ribosome during translation.
-
3' Polyadenylation: A poly(A) tail, a string of adenine nucleotides, is added to the 3' end. This tail also protects the mRNA from degradation and plays a role in its export from the nucleus.
-
Splicing: Eukaryotic genes contain introns, non-coding sequences interspersed within the coding exons. Splicing removes these introns and joins the exons together to create a mature mRNA molecule. This process is mediated by the spliceosome, a complex ribonucleoprotein machine. Alternative splicing, where different combinations of exons can be joined, allows a single gene to produce multiple protein isoforms, significantly increasing proteome diversity. This mechanism is largely absent in prokaryotes, whose genes generally lack introns.
Translational Differences: Beyond the Ribosome
While ribosomes are the primary machinery for translation in both domains, the process differs in several aspects. In prokaryotes, translation initiation often involves a specific ribosome-binding site (Shine-Dalgarno sequence) on the mRNA. This sequence interacts with the 16S rRNA component of the ribosome, facilitating the correct positioning of the ribosome at the translation start codon.
In eukaryotes, the translation initiation process is far more complex. The small ribosomal subunit (40S) binds to the 5' cap of the mRNA, scans the molecule for the start codon (AUG), and then recruits the large ribosomal subunit (60S) to form the functional 80S ribosome. The presence of the 5' cap and the scanning mechanism ensure that translation starts at the correct location. Moreover, eukaryotic translation is highly regulated by various initiation factors, influencing the efficiency of translation initiation.
Regulatory Mechanisms: A Symphony of Control
Gene expression regulation is a crucial aspect of cellular function, allowing organisms to adapt to changing environmental conditions and maintain homeostasis. The regulatory mechanisms are significantly more intricate in eukaryotes.
Prokaryotic Regulation: Operons and Environmental Signals
Prokaryotes primarily regulate gene expression at the transcriptional level through operons. An operon is a cluster of genes transcribed as a single mRNA molecule, often encoding proteins involved in a common metabolic pathway. The expression of operons is controlled by regulatory proteins that bind to specific DNA sequences near the promoter, either activating or repressing transcription. These regulatory proteins often respond to environmental signals, allowing prokaryotes to adjust their gene expression in response to changes in nutrient availability, temperature, or other factors. The lac operon in E. coli, which regulates lactose metabolism, is a classic example of this type of regulation.
Eukaryotic Regulation: A Multi-Layered Approach
Eukaryotic gene regulation is a multi-layered process, encompassing transcriptional, post-transcriptional, translational, and post-translational levels.
Transcriptional Regulation: Eukaryotic promoters are more complex than prokaryotic promoters and involve interactions with a variety of transcription factors, regulatory proteins that bind to specific DNA sequences and influence the rate of transcription initiation. Enhancers and silencers, DNA sequences located far from the promoter, can also influence transcription. Chromatin structure, the organization of DNA into nucleosomes, plays a significant role in regulating gene access to the transcriptional machinery. Histone modification, DNA methylation, and chromatin remodeling complexes all contribute to the control of gene expression.
Post-Transcriptional Regulation: RNA processing itself is a critical regulatory point in eukaryotes. Alternative splicing, as mentioned earlier, can generate multiple protein isoforms from a single gene. RNA interference (RNAi), a mechanism involving small interfering RNAs (siRNAs) and microRNAs (miRNAs), can silence gene expression by degrading mRNA or inhibiting translation. mRNA stability and export from the nucleus are also subject to regulation.
Translational Regulation: The initiation of translation is controlled by various initiation factors, which can be regulated by signaling pathways. Translation can also be inhibited by binding of specific proteins to the mRNA or ribosome.
Post-Translational Regulation: Once a protein is synthesized, its activity can be further regulated by post-translational modifications, including phosphorylation, glycosylation, ubiquitination, and proteolytic cleavage. These modifications can alter the protein's function, localization, or stability.
Evolutionary Implications: From Simplicity to Complexity
The differences in gene expression between prokaryotes and eukaryotes reflect their evolutionary history. The simpler, coupled transcription-translation system of prokaryotes is likely an ancestral feature. The evolution of the eukaryotic nucleus and the development of more complex regulatory mechanisms allowed for greater control over gene expression, leading to increased cellular complexity and specialization. The capacity for alternative splicing and extensive post-transcriptional regulation significantly expanded the functional diversity of eukaryotic genomes.
Conclusion: A Dynamic and Fascinating Field
Gene expression is a fundamental process that underlies all aspects of life. The differences in gene expression mechanisms between prokaryotes and eukaryotes highlight the remarkable diversity of life and the evolutionary pressures that have shaped the genetic systems of these two major domains. Ongoing research continues to unravel the intricate details of gene expression regulation, revealing new layers of complexity and providing deeper insights into the fundamental mechanisms that drive cellular function and development. The study of gene expression remains a vibrant and dynamic field, holding the key to understanding many biological processes, including disease and development. Further exploration into these mechanisms is crucial for the development of new therapeutic interventions and biotechnological applications. The continued study of these fascinating differences promises to unveil further secrets of life's intricate machinery.
Latest Posts
Latest Posts
-
Work Done By An Electric Field
Mar 18, 2025
-
How Much Energy To Be At Zero Kinetic Energy
Mar 18, 2025
-
Current As A Function Of Time
Mar 18, 2025
-
Ode To Billy Joe Lyrics Meaning
Mar 18, 2025
-
Where Does The Light Independent Reaction Take Place
Mar 18, 2025
Related Post
Thank you for visiting our website which covers about Gene Expression In Eukaryotes Vs Prokaryotes . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
