Design Primers For Site Directed Mutagenesis
Muz Play
Mar 25, 2025 · 7 min read
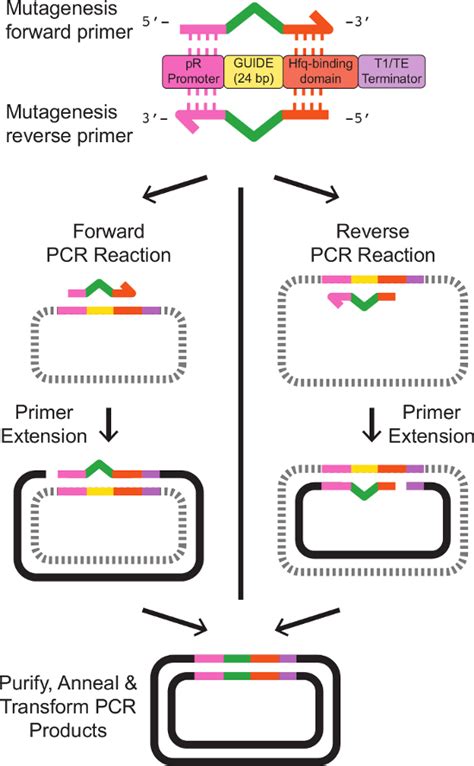
Table of Contents
Design Primers for Site-Directed Mutagenesis: A Comprehensive Guide
Site-directed mutagenesis (SDM) is a powerful molecular biology technique used to introduce specific, targeted changes into a DNA sequence. This allows researchers to study the effects of these changes on gene function, protein structure, and ultimately, the organism's phenotype. A crucial element of successful SDM is the careful design of the primers used in the process. This article provides a comprehensive guide to primer design for SDM, covering key considerations, best practices, and potential pitfalls.
Understanding the Principles of Site-Directed Mutagenesis
Before delving into primer design, let's briefly review the fundamental principles of SDM. The most common method employs PCR to amplify a plasmid containing the target gene. The PCR reaction uses two primers: one that is complementary to the sequence upstream of the desired mutation and another complementary to the sequence downstream, but both containing the desired mutation within their sequence. The amplified plasmid, now containing the mutation, is then transformed into competent cells, replacing the original plasmid.
Several methods exist for SDM, including:
- Traditional PCR-based SDM: This classic approach utilizes two primers, each containing the desired mutation at its 3' end.
- QuikChange® mutagenesis: A commercially available method utilizing a single, circular DNA template and two complementary primers. This method is often favored for its simplicity and high efficiency.
- Overlap extension PCR: This technique employs two separate PCR reactions with overlapping primers containing the desired mutation. The products of these reactions are then combined in a third PCR reaction to generate the full-length mutated gene.
Regardless of the chosen method, effective primer design remains critical for the success of SDM.
Key Considerations for Primer Design in Site-Directed Mutagenesis
Designing primers for SDM requires careful consideration of several factors to ensure high efficiency and accuracy:
1. Primer Length: The Goldilocks Zone
Primer length typically ranges from 25 to 45 base pairs (bp). Primers that are too short may lead to non-specific binding and reduced efficiency, while primers that are too long can result in increased cost and reduced PCR yield. A length within the optimal range generally provides a good balance between specificity and efficiency.
2. Melting Temperature (Tm): Achieving Optimal Annealing
The melting temperature (Tm) is the temperature at which half of the DNA duplex separates into single strands. Ideally, the Tm of both primers should be similar and within the range of 72-78°C. This ensures efficient annealing during PCR. Using online tools, many factors are considered to calculate Tm, such as primer length, GC content, and salt concentration. Inconsistent Tm values between the primers can significantly reduce PCR efficiency.
3. GC Content: Striking a Balance
The GC content, the percentage of guanine and cytosine bases in the primer, should ideally be between 40% and 60%. A higher GC content increases the stability of the primer-template duplex, while a lower GC content can lead to less stable binding and reduced PCR efficiency. Extreme GC content can lead to secondary structures.
4. Self-Complementarity and Hairpin Formation: Avoiding Unwanted Interactions
Primers should be designed to avoid self-complementarity and hairpin formation. These structures can hinder primer annealing and reduce PCR efficiency. Online tools can be used to check for these potential issues, which should be addressed by modifying the primer sequence if necessary.
5. 3'-End Stability: Crucial for Efficient Extension
The 3'-end of the primer is crucial for polymerase extension. It's essential to ensure that the 3'-end of the primer has a high degree of complementarity to the template DNA, with no mismatches or wobbles. Mismatches at the 3'-end can significantly reduce the efficiency of the PCR reaction.
6. Mutation Site Placement: Precisely Locating the Change
The desired mutation should be centrally located within the primer sequence. This generally increases the likelihood of the mutation being incorporated during PCR. Placing the mutation too close to the 5' or 3' end can hinder primer annealing or extension.
7. Primer Design Software: Leveraging Computational Tools
Several software tools are available for primer design, including Primer3, IDT's PrimerQuest, and other commercially available software. These tools can help optimize primer design by considering all the parameters discussed above. Many of these tools are free to use. Remember to check all aspects of the primer before proceeding to synthesis.
Advanced Primer Design Considerations
Beyond the fundamental principles, certain advanced considerations enhance SDM success:
8. Minimizing Mismatches: Ensuring Specificity
Even slight mismatches between the primer and template DNA can decrease PCR efficiency. Careful attention to base pairing is crucial, particularly near the 3' end.
9. Avoiding Repeats: Preventing Secondary Structure Formation
Repeated sequences within the primer can lead to hairpin formation and reduce efficiency. Software tools can detect and help avoid these sequences.
10. Choosing Appropriate Polymerase: Matching Enzyme to the Task
The choice of polymerase significantly impacts SDM efficiency. High-fidelity polymerases, known for their accuracy, are generally preferred to minimize errors during PCR. Selecting a polymerase appropriate for the chosen method of SDM is crucial.
11. Optimizing PCR Conditions: Fine-Tuning the Reaction
While primer design is key, successful SDM also relies on optimal PCR conditions, including annealing temperature, extension time, and the number of cycles. These parameters may need optimization depending on the primer sequences and template DNA.
12. Verification of the Mutation: Confirming Successful Modification
After SDM, it is crucial to verify the successful incorporation of the desired mutation. Sequencing of the modified plasmid is the gold standard for confirming the mutation and the absence of unintended mutations.
Troubleshooting Common Issues in SDM
Even with careful primer design, challenges may arise. Here are some common issues and potential solutions:
- Low PCR yield: This can be due to several factors including poor primer design (low Tm, inappropriate length, mismatches), suboptimal PCR conditions, or template DNA quality.
- Non-specific amplification: This is often caused by primers with low specificity or suboptimal PCR conditions, such as high annealing temperature.
- No amplification: Check for primer dimer formation, hairpin structures, low primer concentration, or inadequate template DNA.
- Incorrect mutation: Verify primer sequences and PCR conditions, and consider re-designing the primers or performing sequencing.
Careful troubleshooting, often iterative, is a key part of successful SDM. Often, re-designing the primers is the answer to many problems.
Examples of Primer Design for Different Mutation Types
Let's illustrate primer design for various mutation types. These are simplified examples and should be further optimized using the guidelines and tools previously mentioned. Always verify the sequences against your target gene.
Example 1: Point Mutation (C to T)
Let's assume the target sequence is: 5'-ATGCGTACGATCGTAGCTAGCT-3' and we want to change the central 'C' to a 'T'.
A possible primer pair could be:
- Forward Primer: 5'-ATGCGTTACGATCGTAGCTAGCT-3'
- Reverse Primer: 5'-AGCTAGCTAGCGATCGCAT-3' (Note: reverse complement)
Example 2: Insertion Mutation
If we wish to insert 'AA' into the same target sequence after the first 'G':
- Forward Primer: 5'-ATGCGTAAACGATCGTAGCTAGCT-3'
- Reverse Primer: 5'-AGCTAGCTACCGATCGCAT-3' (Note: reverse complement)
Example 3: Deletion Mutation
To delete 'CG' from the same sequence after the first 'G':
- Forward Primer: 5'-ATGCGT**ACGATCGTAGCTAGCT-3'
- Reverse Primer: 5'-AGCTAGCTA**CGATCGCAT-3' (Note: reverse complement)
These examples showcase how minor alterations in primer sequences can introduce various mutations. However, always remember to use proper primer design software to verify these sequences and generate optimal primers.
Conclusion: Mastering Primer Design for SDM Success
Site-directed mutagenesis is a powerful tool for studying gene function. Success relies heavily on meticulously designed primers. By carefully considering primer length, Tm, GC content, self-complementarity, 3'-end stability, mutation placement, and utilizing primer design software, researchers can significantly enhance the efficiency and accuracy of their SDM experiments. Remember, thorough verification steps, including DNA sequencing, are essential to confirm the successful introduction of the intended mutation and the absence of unintended alterations. Through understanding these principles and troubleshooting techniques, researchers can effectively utilize SDM to unravel the complexities of gene function and protein structure.
Latest Posts
Latest Posts
-
Political Results Of The Industrial Revolution
Mar 28, 2025
-
Chemical Reactions Form Or Break Between Atoms Ions Or Molecules
Mar 28, 2025
-
Atomic Structure And The Periodic Table
Mar 28, 2025
-
How To Calculate Cumulative Frequency In Excel
Mar 28, 2025
-
Is A Circle Graph A Function
Mar 28, 2025
Related Post
Thank you for visiting our website which covers about Design Primers For Site Directed Mutagenesis . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
