Does Rna Processing Occur In Prokaryotes
Muz Play
Mar 19, 2025 · 5 min read
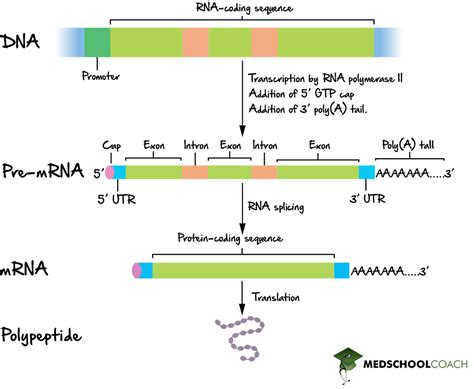
Table of Contents
Does RNA Processing Occur in Prokaryotes? A Deep Dive into Transcription and Translation
The central dogma of molecular biology dictates that genetic information flows from DNA to RNA to protein. While this process is fundamentally conserved across all life forms, the complexity of each step varies significantly between prokaryotes and eukaryotes. A key difference lies in RNA processing. Eukaryotic RNA undergoes extensive processing before translation, including splicing, capping, and polyadenylation. But does RNA processing occur in prokaryotes? The short answer is: yes, but to a much lesser extent than in eukaryotes. This article will delve into the nuances of prokaryotic RNA processing, exploring the similarities and differences with eukaryotic systems and clarifying common misconceptions.
The Core Difference: Coupled Transcription and Translation
The most significant factor influencing the extent of RNA processing in prokaryotes is the coupling of transcription and translation. In eukaryotes, these processes are spatially and temporally separated: transcription occurs in the nucleus, and translation takes place in the cytoplasm. This separation allows for extensive RNA processing to occur between the two steps. In prokaryotes, however, transcription and translation occur simultaneously in the cytoplasm. As the mRNA molecule is being transcribed, ribosomes bind to its 5' end and begin translation.
This close coupling significantly reduces the need for extensive RNA processing. There's simply less time and opportunity for modifications to occur before the mRNA is translated into protein. While some processing does take place, it’s far less elaborate and diverse than in eukaryotes.
Types of RNA Processing in Prokaryotes
While prokaryotes lack the complex machinery for splicing, capping, and polyadenylation seen in eukaryotes, several processing events do occur, albeit in a simpler form:
1. Transcriptional Attenuation: A Regulatory Mechanism
Attenuation is a form of transcriptional regulation where transcription of a gene is prematurely terminated. It's crucial for controlling the expression of genes involved in amino acid biosynthesis. When levels of a specific amino acid are high, ribosomes translate the mRNA quickly, preventing the formation of a secondary structure that would normally signal termination. Conversely, when amino acid levels are low, translation slows down, allowing the termination structure to form, halting transcription. This process is a form of RNA processing because it directly affects the final length and therefore the functionality of the transcribed RNA molecule.
2. Ribosomal RNA (rRNA) and Transfer RNA (tRNA) Processing
Prokaryotic rRNA and tRNA molecules also undergo processing, although this is simpler than their eukaryotic counterparts. These molecules are transcribed as large precursor molecules that subsequently undergo cleavage and modification.
- Cleavage: Specific endoribonucleases cleave the precursor molecules at precise sites to generate mature rRNA and tRNA.
- Base Modification: Several bases within the rRNA and tRNA molecules undergo chemical modifications, such as methylation or pseudouridylation. These modifications are essential for the proper function of these molecules in translation.
These processing steps are crucial for ensuring the functionality of the ribosome and the accuracy of translation. They are considered forms of RNA processing because they involve the post-transcriptional alteration of the primary RNA transcript to yield functional molecules.
3. mRNA Processing: A Simpler Affair
Prokaryotic mRNA processing is significantly less complex than in eukaryotes. While there's no equivalent to splicing, capping, or polyadenylation, several subtle processing events can influence the stability and translation efficiency of mRNA:
- Leader Sequence Modifications: The leader sequence (the region upstream of the start codon) can influence ribosome binding and translation initiation. Some leader sequences undergo structural changes or modifications that impact their ability to bind ribosomes.
- mRNA Degradation: Prokaryotic mRNA molecules are typically less stable than their eukaryotic counterparts, with shorter half-lives. This reflects the rapid turnover of proteins in prokaryotes. The degradation of mRNA is itself a form of RNA processing, removing it from translational machinery.
- Riboswitches: Certain mRNA molecules contain regulatory regions known as riboswitches. These regions bind to specific metabolites, causing structural changes in the mRNA and affecting its translation. These structures act as direct regulators of gene expression through structural changes in the RNA itself.
Comparing Prokaryotic and Eukaryotic RNA Processing: A Summary Table
| Feature | Prokaryotes | Eukaryotes |
|---|---|---|
| Transcription | Coupled with translation in cytoplasm | Occurs in nucleus, separated from translation |
| Splicing | Absent | Present, removes introns |
| 5' Capping | Absent | Present, protects mRNA and enhances translation |
| 3' Polyadenylation | Absent | Present, protects mRNA and signals for translation |
| rRNA Processing | Cleavage and base modification | Extensive cleavage, base modification, and methylation |
| tRNA Processing | Cleavage and base modification | Extensive cleavage, base modification, and splicing |
| mRNA Processing | Limited; attenuation, leader sequence modification, riboswitches and degradation | Extensive; splicing, capping, polyadenylation, editing |
| mRNA Stability | Generally shorter half-life | Generally longer half-life |
The Importance of Understanding Prokaryotic RNA Processing
Understanding the nuances of RNA processing in prokaryotes is crucial for several reasons:
- Antibiotic Development: Many antibiotics target prokaryotic ribosomes or other components of the protein synthesis machinery. A detailed understanding of prokaryotic RNA processing could aid in the development of new antibiotics targeting these processes.
- Genetic Engineering: Manipulating gene expression in prokaryotes is essential for biotechnology applications, such as the production of recombinant proteins. Knowing the intricacies of prokaryotic RNA processing can help to optimize these processes.
- Understanding Bacterial Pathogenesis: Understanding how bacteria regulate gene expression, including RNA processing, is crucial to understand the mechanisms of bacterial pathogenicity and develop new treatments.
- Evolutionary Biology: Comparing RNA processing in prokaryotes and eukaryotes provides valuable insights into the evolution of gene regulation and the development of cellular complexity.
Conclusion: Subtle but Significant
While prokaryotes don't exhibit the elaborate RNA processing machinery of eukaryotes, several important processing events do occur. These processes are crucial for regulating gene expression, ensuring the proper function of ribosomes, and maintaining the stability of mRNA molecules. The absence of extensive processing reflects the coupled nature of transcription and translation in prokaryotes, which simplifies the need for intermediate steps. However, these simpler forms of RNA processing play vital roles in cellular function and are worthy of careful study. Further research will undoubtedly illuminate more subtle aspects of prokaryotic RNA processing, enhancing our understanding of bacterial biology and offering new opportunities for biotechnological and therapeutic interventions.
Latest Posts
Latest Posts
-
How To Find The Matrix Of A Linear Transformation
Mar 19, 2025
-
Find The Projection Of U Onto V
Mar 19, 2025
-
How Many Valence Electrons In Iron
Mar 19, 2025
-
Variation Of Parameters Method Differential Equations
Mar 19, 2025
-
How Is Redshift Related To Distance Equation
Mar 19, 2025
Related Post
Thank you for visiting our website which covers about Does Rna Processing Occur In Prokaryotes . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
