Which Molecules In Eukaryotic Cells Regulate Gene Expression
Muz Play
Apr 01, 2025 · 7 min read
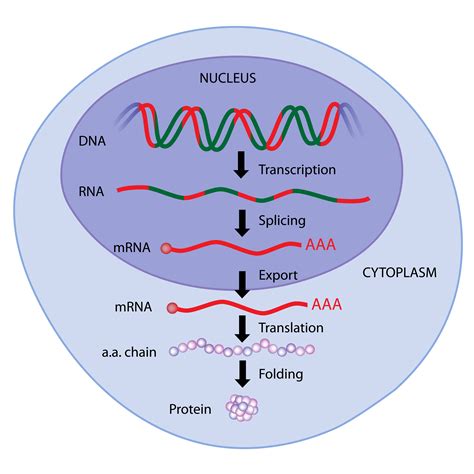
Table of Contents
Which Molecules in Eukaryotic Cells Regulate Gene Expression?
Gene expression, the intricate process of converting genetic information encoded in DNA into functional gene products (proteins or functional RNAs), is tightly regulated in eukaryotic cells. This regulation ensures that genes are expressed only when and where needed, maintaining cellular homeostasis and enabling specialized functions within multicellular organisms. A complex interplay of numerous molecules orchestrates this precise control, operating at various stages from DNA accessibility to protein degradation. Understanding these regulatory molecules is crucial to grasping the complexity of life and addressing various diseases arising from gene expression dysregulation.
DNA Accessibility and Chromatin Remodeling: The Foundation of Gene Regulation
The first and often rate-limiting step in gene expression is access to the DNA itself. Eukaryotic DNA is packaged into chromatin, a complex structure of DNA wound around histone proteins. The degree of chromatin compaction significantly influences the accessibility of genes to the transcriptional machinery. Several molecules play critical roles in remodeling chromatin to either promote or repress gene expression:
1. Histone Modifying Enzymes: Writing and Erasing the Histone Code
Histones are subject to various post-translational modifications, including acetylation, methylation, phosphorylation, and ubiquitination. These modifications, collectively known as the "histone code," alter chromatin structure and influence gene expression.
-
Histone acetyltransferases (HATs): These enzymes add acetyl groups to lysine residues on histone tails, neutralizing their positive charge. This weakens the interaction between histones and DNA, leading to chromatin decondensation and increased gene accessibility – generally associated with transcriptional activation.
-
Histone deacetylases (HDACs): Conversely, HDACs remove acetyl groups, leading to chromatin condensation and transcriptional repression. HDAC inhibitors are currently being explored as therapeutic agents in various cancers due to their ability to reactivate tumor suppressor genes silenced by HDAC-mediated repression.
-
Histone methyltransferases (HMTs): These enzymes add methyl groups to lysine and arginine residues on histone tails. Methylation can either activate or repress transcription depending on the specific residue modified and the number of methyl groups added. For example, H3K4 methylation is generally associated with activation, while H3K9 methylation is associated with repression.
-
Histone demethylases (HDMs): These enzymes remove methyl groups from histones, counteracting the effects of HMTs and contributing to dynamic regulation of gene expression.
2. Chromatin Remodelers: Reshaping the Chromatin Landscape
Chromatin remodeling complexes are large multi-protein machines that use ATP hydrolysis to alter nucleosome positioning and spacing. This dynamic repositioning of nucleosomes can expose or mask promoter regions, influencing the ability of transcription factors to bind and initiate transcription. These complexes are crucial for both activation and repression of gene expression, contributing to the plasticity of the genome. Examples include the SWI/SNF and ISWI families of complexes.
Transcriptional Regulation: Orchestrating the Initiation of Transcription
Once the chromatin is remodeled to make the gene accessible, the actual process of transcription can begin. This stage involves the recruitment of a complex array of molecules to the promoter region of the gene:
1. Transcription Factors: The Master Regulators
Transcription factors (TFs) are proteins that bind to specific DNA sequences (cis-regulatory elements) within the promoter and enhancer regions of genes. They act as molecular switches, either activating or repressing transcription. They achieve this through various mechanisms:
-
Activating TFs: Recruit components of the basal transcriptional machinery, including RNA polymerase II and general transcription factors (GTFs), to the promoter. Many activating TFs possess activation domains that interact with coactivators, further enhancing transcriptional activity.
-
Repressing TFs: Compete with activating TFs for binding to DNA, block the binding of RNA polymerase, or recruit corepressors that inhibit transcription. Some repressor TFs can also recruit HDACs, leading to chromatin condensation and transcriptional silencing.
2. Coactivators and Corepressors: Modulating Transcriptional Activity
Coactivators and corepressors are proteins that interact with TFs to enhance or suppress transcription, respectively. They don't directly bind to DNA but play crucial roles in modulating the activity of TFs. Some coactivators possess histone acetyltransferase activity, while others act as bridging molecules linking TFs to the basal transcriptional machinery. Corepressors often recruit HDACs or other chromatin-modifying enzymes.
3. RNA Polymerase II and General Transcription Factors: The Transcriptional Machinery
RNA polymerase II is the enzyme responsible for transcribing protein-coding genes. It cannot initiate transcription on its own and requires the assistance of general transcription factors (GTFs), a set of proteins that assemble at the promoter to form the pre-initiation complex. The assembly and activity of this complex are tightly regulated by TFs and coactivators/corepressors.
Post-Transcriptional Regulation: Fine-Tuning Gene Expression
Even after transcription is initiated, gene expression can be further regulated at multiple post-transcriptional levels:
1. RNA Processing: Splicing, Capping, and Polyadenylation
The primary RNA transcript undergoes several processing steps before it can be translated into protein. These steps include:
-
Splicing: Removal of introns and joining of exons. Alternative splicing allows for the production of multiple protein isoforms from a single gene, expanding proteome diversity. Spliceosome components and regulatory splicing factors contribute to the complexity of this process.
-
Capping: Addition of a 5' cap, which protects the mRNA from degradation and promotes translation initiation.
-
Polyadenylation: Addition of a poly(A) tail to the 3' end, enhancing mRNA stability and translation efficiency.
2. RNA Stability and Degradation: Controlling mRNA Lifespan
The lifespan of an mRNA molecule significantly impacts the amount of protein produced. mRNA stability is influenced by various factors, including the presence of specific sequences in the 3' untranslated region (UTR) and the binding of RNA-binding proteins (RBPs). RBPs can either stabilize or destabilize mRNA, influencing its half-life. RNA degradation pathways, involving exonucleases and endonucleases, also play a critical role in controlling mRNA levels.
3. RNA Interference (RNAi): Silencing Gene Expression through RNA Molecules
RNAi is a mechanism by which small RNA molecules, such as microRNAs (miRNAs) and short interfering RNAs (siRNAs), can regulate gene expression. These small RNAs bind to complementary sequences in target mRNAs, leading to either mRNA degradation or translational repression. The RNA-induced silencing complex (RISC) is a key player in RNAi, mediating the interaction between small RNAs and target mRNAs.
4. Translational Regulation: Controlling Protein Synthesis
The initiation and efficiency of protein synthesis can be regulated by various factors, including:
-
Initiation factors: Proteins involved in assembling the ribosome on the mRNA and initiating translation. The availability and activity of these factors can be regulated.
-
RNA-binding proteins (RBPs): Can bind to mRNAs and either promote or inhibit translation initiation.
-
Phosphorylation of translation initiation factors: Phosphorylation can either activate or inhibit initiation factors, affecting the rate of translation.
Post-Translational Regulation: Fine-Tuning Protein Function
Even after a protein is synthesized, its activity and lifespan can be regulated:
1. Protein Degradation: Controlling Protein Levels
Proteins are continuously degraded through various pathways, such as the ubiquitin-proteasome system (UPS) and autophagy. The UPS targets proteins for degradation by attaching ubiquitin chains, marking them for recognition by the proteasome, a large protein complex that degrades ubiquitinated proteins. Autophagy is another process involved in the degradation of damaged or misfolded proteins.
2. Protein Modification: Altering Protein Activity
Proteins can undergo various post-translational modifications, including phosphorylation, glycosylation, acetylation, and ubiquitination. These modifications can alter protein activity, localization, or stability. Kinases and phosphatases regulate phosphorylation, while other enzymes catalyze other types of modifications.
Conclusion: A Complex Orchestration of Molecular Players
Regulation of gene expression in eukaryotic cells is a remarkably complex process, involving a coordinated interplay of numerous molecules operating at multiple levels. From chromatin remodeling to protein degradation, each stage provides opportunities for fine-tuning gene expression to meet the ever-changing needs of the cell. Understanding these regulatory mechanisms is not only fundamental to basic biology but also crucial for developing novel therapeutic strategies for treating diseases arising from gene expression dysregulation, including cancer, developmental disorders, and neurological diseases. Further research into the intricacies of gene regulation continues to unveil the amazing complexity and precision of cellular control.
Latest Posts
Latest Posts
-
Solving Inequalities With Absolute Value Worksheet
Apr 02, 2025
-
What Happens To Volume When Temperature Increases
Apr 02, 2025
-
What Is The Difference Between Ethnic And Religious Groups
Apr 02, 2025
-
Explain The Difference Between An Autotroph And A Heterotroph
Apr 02, 2025
-
Is Magnesium A Metal Nonmetal Or Metalloid
Apr 02, 2025
Related Post
Thank you for visiting our website which covers about Which Molecules In Eukaryotic Cells Regulate Gene Expression . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
