Differences In Gene Expression Between Prokaryotes And Eukaryotes
Muz Play
Mar 18, 2025 · 6 min read
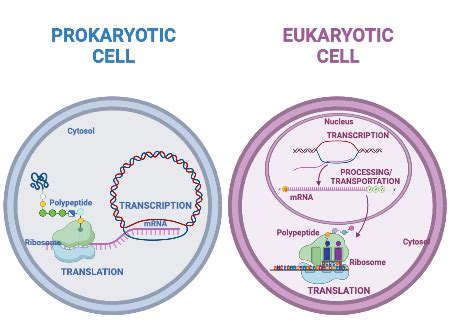
Table of Contents
The Great Divide: Unveiling the Differences in Gene Expression Between Prokaryotes and Eukaryotes
Gene expression, the intricate process by which information encoded within genes is translated into functional proteins, is a fundamental aspect of life. While the underlying principle—DNA to RNA to protein—remains consistent across all living organisms, the mechanisms and complexities involved exhibit significant differences between prokaryotes (bacteria and archaea) and eukaryotes (plants, animals, fungi, and protists). Understanding these differences is crucial for comprehending the vast diversity of life and holds immense implications for various fields, including medicine, biotechnology, and evolutionary biology.
Fundamental Differences: A Tale of Two Cells
The most striking difference lies in the cellular organization of prokaryotes and eukaryotes. Prokaryotic cells lack a defined nucleus and other membrane-bound organelles, resulting in a simpler cellular architecture. Eukaryotic cells, in contrast, boast a complex internal structure, with DNA housed within a membrane-enclosed nucleus and numerous other organelles, each performing specialized functions. This fundamental structural disparity significantly impacts how gene expression unfolds.
1. Transcriptional Regulation: A Complex Orchestra
Prokaryotic Transcription: In prokaryotes, transcription and translation are coupled—they occur simultaneously in the cytoplasm. This allows for rapid response to environmental changes. Transcriptional regulation is primarily achieved through operons, clusters of genes under the control of a single promoter. Repressors and activators, proteins that bind to specific DNA sequences near the promoter, modulate transcription levels. The lac operon in E. coli, a classic example, regulates lactose metabolism based on the availability of lactose and glucose.
Eukaryotic Transcription: Eukaryotic transcription is a far more intricate process, spatially and temporally separated from translation. It takes place within the nucleus, involving numerous transcription factors that interact with the promoter and enhancer regions of the gene. Chromatin remodeling, the alteration of chromatin structure to make DNA accessible to the transcriptional machinery, plays a critical role. Eukaryotic genes also possess introns (non-coding sequences) interspersed within exons (coding sequences), requiring RNA splicing to remove introns and produce mature mRNA. The complexity is further amplified by alternative splicing, which can generate multiple protein isoforms from a single gene.
Key Differences Summarized:
| Feature | Prokaryotes | Eukaryotes |
|---|---|---|
| Location | Cytoplasm | Nucleus |
| Coupling | Transcription and translation are coupled | Transcription and translation are uncoupled |
| Regulation | Primarily operons, repressors, activators | Complex interplay of transcription factors, chromatin remodeling |
| RNA processing | Minimal processing | Extensive processing (splicing, capping, polyadenylation) |
2. RNA Processing: Maturation and Modification
The journey of mRNA from nascent transcript to mature, translatable molecule differs greatly between prokaryotes and eukaryotes.
Prokaryotic RNA Processing: Prokaryotic mRNA undergoes minimal processing. It is typically polycistronic, meaning it encodes multiple proteins from a single transcript. Once transcribed, it can immediately bind to ribosomes and initiate translation.
Eukaryotic RNA Processing: Eukaryotic mRNA undergoes extensive processing within the nucleus before export to the cytoplasm. This involves:
- 5' capping: Addition of a 7-methylguanosine cap to the 5' end, protecting the mRNA from degradation and enhancing ribosome binding.
- Splicing: Removal of introns and ligation of exons to form a continuous coding sequence. Alternative splicing adds layers of complexity, generating protein diversity.
- 3' polyadenylation: Addition of a poly(A) tail to the 3' end, protecting the mRNA from degradation and facilitating export from the nucleus.
This meticulous processing ensures mRNA stability and efficient translation in eukaryotes. The absence of these steps in prokaryotes reflects their faster, simpler gene expression machinery.
3. Translational Regulation: Fine-Tuning Protein Synthesis
Prokaryotic Translation: Prokaryotic translation initiation is relatively straightforward. Ribosomes bind directly to the mRNA's ribosome-binding site (Shine-Dalgarno sequence), initiating protein synthesis.
Eukaryotic Translation: Eukaryotic translation is a more intricate process, involving several initiation factors and the 5' cap structure. The ribosome binds to the 5' cap and scans the mRNA until it encounters the start codon (AUG). Translation can be regulated through various mechanisms, including:
- Initiation factor regulation: Availability and activity of initiation factors can control translation rates.
- RNA interference (RNAi): Small interfering RNAs (siRNAs) and microRNAs (miRNAs) can bind to mRNA, leading to its degradation or translational repression.
- Phosphorylation of initiation factors: Phosphorylation status of initiation factors can modulate their ability to initiate translation.
4. Post-Translational Modifications: Protein Maturation and Function
Prokaryotic Post-Translational Modifications: Post-translational modifications in prokaryotes are relatively less common compared to eukaryotes. Common modifications include protein folding, proteolytic cleavage, and the addition of small molecules like methyl groups.
Eukaryotic Post-Translational Modifications: Eukaryotic proteins undergo a wide range of post-translational modifications, impacting their structure, function, localization, and stability. These include:
- Glycosylation: Addition of sugar moieties.
- Phosphorylation: Addition of phosphate groups.
- Ubiquitination: Addition of ubiquitin, targeting proteins for degradation.
- Acetylation: Addition of acetyl groups.
- Proteolytic cleavage: Removal of amino acid sequences.
These modifications add an extra layer of control over protein activity and contribute to the diversity of eukaryotic cellular functions.
Evolutionary Implications: From Simple to Complex
The differences in gene expression mechanisms between prokaryotes and eukaryotes reflect their evolutionary trajectories. Prokaryotes, with their simpler cellular structure and coupled transcription-translation, evolved strategies for rapid adaptation to fluctuating environments. Eukaryotes, with their complex cellular organization and compartmentalization, developed sophisticated regulatory mechanisms to control gene expression with greater precision and control. This increased complexity allowed for the evolution of multicellular organisms with specialized cell types and intricate developmental processes.
Technological Applications: Harnessing the Differences
The contrasting features of prokaryotic and eukaryotic gene expression are exploited in various biotechnological applications:
- Prokaryotic systems (e.g., E. coli) are widely used for recombinant protein production due to their simplicity, rapid growth, and well-established genetic manipulation techniques.
- Eukaryotic systems (e.g., yeast, mammalian cells) are employed when post-translational modifications are required for proper protein folding and function, making them valuable tools in producing therapeutic proteins.
Conclusion: A Continuing Story
The differences in gene expression between prokaryotes and eukaryotes are a testament to the remarkable diversity of life. While the fundamental principles remain the same, the intricacies of regulation and complexity highlight the evolutionary adaptations required for survival and diversification in diverse environments. Continued research into these mechanisms will undoubtedly unveil further insights into cellular processes, human health, and the development of novel biotechnological applications. Further studies on specific gene families, their regulation, and the interplay between different regulatory elements will deepen our understanding of this fundamental biological process. The comparative study of gene expression across different organisms will continue to illuminate the evolutionary pathways and provide a rich foundation for future breakthroughs in various scientific disciplines. Understanding these differences remains crucial in developing novel therapeutics, advancing biotechnology, and ultimately, gaining a deeper appreciation for the elegance and complexity of life itself.
Latest Posts
Latest Posts
-
How To Write Mass Balance Equations
Mar 18, 2025
-
A Virus That Undergoes Lysogeny Is A An
Mar 18, 2025
-
Where Are Metals Located In The Periodic Table
Mar 18, 2025
-
Gay Lussac Law Of Combining Volume
Mar 18, 2025
-
How To Analysis Mass Spectrometry Data
Mar 18, 2025
Related Post
Thank you for visiting our website which covers about Differences In Gene Expression Between Prokaryotes And Eukaryotes . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
