Lock And Key Vs Induced Fit
Muz Play
Mar 16, 2025 · 6 min read
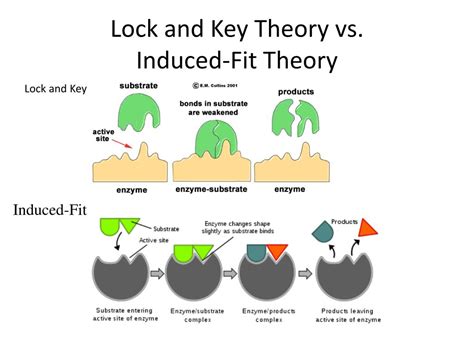
Table of Contents
Lock and Key vs. Induced Fit: A Deep Dive into Enzyme-Substrate Interactions
The interaction between enzymes and their substrates is a fundamental process in biochemistry, driving countless reactions essential for life. Two prominent models have historically been used to explain this interaction: the lock and key model and the induced fit model. While the lock and key model provides a simplified initial understanding, the induced fit model offers a more nuanced and accurate representation of the dynamic nature of enzyme-substrate binding. This article will delve into both models, comparing and contrasting their features, exploring their limitations, and highlighting the current understanding of enzyme-substrate interactions.
The Lock and Key Model: A Simple Analogy
The lock and key model, proposed by Emil Fischer in 1894, likens the enzyme to a lock and the substrate to a key. This model posits that the enzyme possesses a rigid, pre-formed active site with a specific three-dimensional shape complementary to the shape of its substrate. Only the correct substrate, possessing the precise shape and chemical properties, can fit into the active site, initiating the catalytic process. This interaction is often described as a "perfect fit," implying a high degree of specificity and a rigid enzyme structure.
Advantages of the Lock and Key Model:
- Simplicity: The model provides a straightforward and easily understandable analogy, making it a useful introductory concept in biochemistry.
- Specificity: It explains the high degree of specificity observed in many enzyme-substrate interactions, where only certain substrates can bind to a particular enzyme.
- Initial Understanding: It served as a valuable foundational model that spurred further research into enzyme kinetics and mechanism.
Limitations of the Lock and Key Model:
- Rigidity: The model assumes a completely rigid enzyme structure, which is not entirely accurate. Enzymes are flexible macromolecules capable of conformational changes.
- Lack of Dynamic Interaction: It fails to account for the dynamic interactions and conformational changes that occur during substrate binding and catalysis.
- Limited Applicability: While effective for some enzyme-substrate pairs, it cannot explain the interactions of enzymes with multiple substrates or the induced fit phenomena observed in many enzymatic reactions.
The Induced Fit Model: A More Realistic Representation
The induced fit model, proposed by Daniel Koshland in 1958, refines the lock and key model by incorporating the flexibility of enzymes. This model suggests that the enzyme's active site is not a rigid, pre-formed structure but rather a flexible site that adapts its shape to accommodate the substrate upon binding. The substrate's binding induces a conformational change in the enzyme, optimizing the interaction and facilitating catalysis. This dynamic interaction involves both the enzyme and the substrate undergoing conformational changes to achieve the optimal transition state for the reaction.
Advantages of the Induced Fit Model:
- Flexibility: It acknowledges the dynamic nature of enzyme-substrate interactions and the flexibility of enzyme structures.
- Enhanced Specificity: The conformational changes induced upon substrate binding enhance specificity by optimizing the active site for catalysis.
- Explanation of Multiple Substrates: It explains how enzymes can interact with multiple substrates with varying structures, often exhibiting broader substrate specificity.
- Transition State Stabilization: The induced fit model better explains how enzymes stabilize the transition state of the reaction, lowering the activation energy and increasing the reaction rate.
The Role of Non-Covalent Interactions in Induced Fit
The conformational changes driving induced fit are primarily mediated by various non-covalent interactions between the enzyme and the substrate. These interactions include:
- Hydrogen Bonds: Weak but numerous hydrogen bonds contribute to the overall binding energy and shape complementarity.
- Ionic Interactions: Electrostatic attractions between charged amino acid residues on the enzyme and the substrate.
- Hydrophobic Interactions: The tendency of nonpolar groups to cluster together, driven by the hydrophobic effect, plays a significant role in substrate binding and orientation.
- Van der Waals Forces: Weak, short-range attractive forces contributing to overall binding affinity.
The interplay of these non-covalent interactions creates a delicate balance that drives the conformational adjustments in both the enzyme and the substrate, leading to the formation of the enzyme-substrate complex.
Experimental Evidence Supporting Induced Fit
Numerous experimental techniques provide compelling evidence for the induced fit model. These include:
- X-ray crystallography: This technique allows for the determination of the three-dimensional structure of enzymes, both in the absence and presence of substrates. The comparison of these structures often reveals significant conformational changes upon substrate binding, supporting the induced fit mechanism.
- Nuclear magnetic resonance (NMR) spectroscopy: NMR provides information about the dynamic interactions between enzymes and substrates, revealing conformational changes during the binding process.
- Kinetic studies: Kinetic experiments examining the rate of enzymatic reactions can provide insights into the binding mechanism. Observations supporting induced fit include the presence of multiple binding steps and conformational changes affecting the rate of catalysis.
- Site-directed mutagenesis: By altering specific amino acid residues in the enzyme's active site, researchers can probe the role of individual residues in substrate binding and catalysis. Mutations affecting conformational changes often lead to altered enzymatic activity, further validating the induced fit model.
Comparing the Lock and Key and Induced Fit Models
| Feature | Lock and Key Model | Induced Fit Model |
|---|---|---|
| Enzyme Active Site | Rigid, pre-formed | Flexible, adapts to substrate upon binding |
| Substrate Binding | "Perfect fit," precise complementarity | Conformational changes in both enzyme and substrate |
| Specificity | High, determined by pre-formed shape | High, enhanced by induced fit |
| Dynamic Interaction | No significant conformational changes | Significant conformational changes |
| Applicability | Limited, applies to some enzymes | More widely applicable, reflects reality more accurately |
Beyond the Models: A More Comprehensive View
While the induced fit model provides a more accurate representation of enzyme-substrate interactions than the lock and key model, it is also a simplification. The reality is often far more complex, involving a multitude of factors beyond simple shape complementarity.
These additional factors include:
- Solvent effects: The surrounding aqueous environment plays a significant role in shaping the enzyme's structure and its interaction with the substrate.
- Allosteric regulation: Enzymes can be regulated by molecules binding to sites other than the active site, inducing conformational changes that affect substrate binding and catalysis.
- Protein dynamics: The inherent flexibility of enzymes is far more complex than simple induced fit. Internal motions and fluctuations within the enzyme can contribute to substrate binding and catalysis.
- Enzyme-substrate interactions beyond the active site: Interactions outside the active site can also contribute to substrate binding and orientation, influencing catalysis.
Conclusion: The Dynamic Dance of Enzymes and Substrates
The lock and key and induced fit models represent significant steps in understanding enzyme-substrate interactions. While the lock and key model offers a simplified initial understanding, the induced fit model provides a more accurate and comprehensive view of the dynamic nature of this fundamental biological process. It emphasizes the flexibility of enzymes and the crucial role of conformational changes in achieving efficient catalysis. However, recognizing the limitations of even the induced fit model, modern research increasingly appreciates the intricate and multifaceted nature of enzyme-substrate interactions, involving factors extending beyond simple shape complementarity and conformational changes. This dynamic interplay of forces ultimately underpins the remarkable efficiency and specificity of enzymatic reactions essential to life itself. Further research continues to refine our understanding of these intricate interactions, leveraging advanced techniques to unveil the complexities of this essential biological process. The field remains a fertile ground for ongoing investigation and innovation.
Latest Posts
Latest Posts
-
A Mixture In Which The Composition Is Uniform Throughout
Mar 16, 2025
-
How To Break The Nitrogen Off An Imine Mechanism
Mar 16, 2025
-
Changes Color At The Endpoint Of A Titration
Mar 16, 2025
-
An Element That Conducts Heat And Electricity Poorly
Mar 16, 2025
-
Can Their Be Lessd Gpe Than Kpe
Mar 16, 2025
Related Post
Thank you for visiting our website which covers about Lock And Key Vs Induced Fit . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
