Primer Design For Site Directed Mutagenesis
Muz Play
Mar 27, 2025 · 6 min read
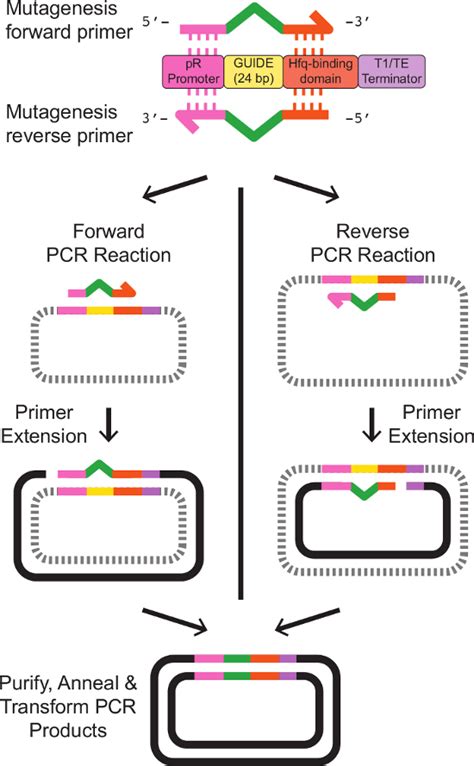
Table of Contents
- Primer Design For Site Directed Mutagenesis
- Table of Contents
- Primer Design for Site-Directed Mutagenesis: A Comprehensive Guide
- Understanding the Basics of Site-Directed Mutagenesis
- Key Considerations for Primer Design
- 1. Primer Length and Melting Temperature (Tm):
- 2. GC Content:
- 3. Primer Self-Complementarity and Hairpin Formation:
- 4. 3' End Stability:
- 5. Mutation Site Incorporation:
- 6. Avoiding Repeat Sequences:
- 7. Specificity Checks:
- Designing Primers for Different Mutation Types
- Point Mutations:
- Insertions and Deletions:
- Multiple Mutations:
- Utilizing Online Primer Design Tools
- Advanced Considerations
- Troubleshooting Common Problems
- Conclusion
- Latest Posts
- Latest Posts
- Related Post
Primer Design for Site-Directed Mutagenesis: A Comprehensive Guide
Site-directed mutagenesis (SDM) is a powerful molecular biology technique used to introduce specific, targeted changes into a DNA sequence. This allows researchers to study the effects of these changes on protein function, structure, and expression. The cornerstone of successful SDM lies in the meticulous design of the mutagenic primers. This article provides a comprehensive guide to primer design for SDM, covering crucial aspects from basic principles to advanced considerations.
Understanding the Basics of Site-Directed Mutagenesis
Before delving into primer design, it's essential to understand the underlying principles of SDM. The most common method utilizes PCR to amplify a plasmid containing the target gene. The PCR reaction includes two primers: one forward and one reverse. Crucially, these primers incorporate the desired mutation(s) within their sequence. After amplification, the parental plasmid is digested using a restriction enzyme (often DpnI, which specifically digests methylated DNA), leaving only the newly synthesized, mutated plasmid. This mutated plasmid is then transformed into competent cells for further analysis.
Key Considerations for Primer Design
Effective primer design is paramount for successful SDM. Several crucial factors influence primer efficacy:
1. Primer Length and Melting Temperature (Tm):
- Optimal Length: Primers are typically 25-45 bases long. Shorter primers may lack specificity, leading to non-specific amplification. Longer primers may suffer from reduced efficiency and increased cost.
- Melting Temperature (Tm): The Tm is the temperature at which 50% of the primer hybridizes to its complementary strand. A well-designed primer usually has a Tm between 72-80°C. Several online tools and equations (such as the nearest neighbor method) can accurately calculate the Tm. Consistency in Tm between the forward and reverse primers is crucial for optimal PCR efficiency.
2. GC Content:
- Ideal Range: The GC content should ideally be between 40-60%. This balance prevents excessive secondary structure formation and ensures stable hybridization. Extremely high or low GC content can negatively impact primer annealing and amplification.
3. Primer Self-Complementarity and Hairpin Formation:
- Avoiding Secondary Structures: Primers should be checked for self-complementarity and hairpin formation. These secondary structures can hinder primer annealing and reduce PCR efficiency. Numerous online tools can predict and analyze these potential problems. Modifying the primer sequence slightly can often mitigate these issues.
4. 3' End Stability:
- Importance of the 3' End: The 3' end of the primer is crucial for polymerase extension. It should be highly complementary to the target template to ensure efficient extension. Avoid mismatches or unstable sequences at the 3' end. This is especially critical for the region surrounding the mutation site.
5. Mutation Site Incorporation:
- Strategic Placement: The mutation(s) should be strategically placed within the primer sequence. A common strategy is to incorporate the mutation near the center of the primer to ensure stable hybridization. The exact position will depend on the specific mutation and the overall primer design. Avoid placing the mutation too close to the 3' end, as this can interfere with polymerase extension.
6. Avoiding Repeat Sequences:
- Minimizing Non-Specific Binding: Primers should avoid long stretches of repetitive sequences. Such sequences increase the chance of non-specific binding to other regions of the genome. This can lead to the amplification of unwanted products.
7. Specificity Checks:
- BLAST Search: Before synthesis, primers should be subjected to a BLAST search against the target genome and other relevant databases. This will help ensure that the primers are specific to the intended target and avoid potential off-target effects.
Designing Primers for Different Mutation Types
The approach to primer design varies slightly depending on the type of mutation being introduced:
Point Mutations:
For point mutations (single base changes), the primer is designed to incorporate the desired base change at the appropriate position. The surrounding sequence should be highly complementary to the template DNA. It’s vital to ensure that the mutation doesn't significantly alter the primer's Tm or create secondary structures.
Insertions and Deletions:
Introducing insertions or deletions requires a more nuanced approach. The primer must contain the intended insertion or deletion, and the flanking sequences must be carefully chosen to maintain primer stability and ensure efficient amplification. It's often beneficial to design primers with longer overhangs to accommodate larger insertions or deletions.
Multiple Mutations:
Introducing multiple mutations simultaneously involves incorporating all desired changes into the primer sequences. This requires careful consideration of primer length, Tm, and potential secondary structures. It might be more efficient to introduce multiple mutations sequentially rather than in a single step.
Utilizing Online Primer Design Tools
Several online tools greatly simplify primer design for SDM. These tools offer various features such as:
- Tm calculation: Accurate calculation of the melting temperature.
- Secondary structure prediction: Identification of potential self-complementarity or hairpin formation.
- GC content analysis: Assessment of the GC content.
- BLAST search: Checking for specificity.
While these tools are invaluable, it's essential to understand the underlying principles of primer design and critically evaluate the results generated by these programs. Don't solely rely on automated tools; always manually check the design parameters.
Advanced Considerations
- Using different polymerases: The choice of polymerase significantly impacts PCR efficiency. Some polymerases are better suited for SDM than others. Consider high-fidelity polymerases to minimize errors during amplification.
- Primer optimization: If the initial primers yield unsatisfactory results, optimization might be necessary. This could involve adjusting primer length, Tm, or GC content.
- Colony PCR screening: Verify the presence of the mutation in the resulting colonies using colony PCR. This is a crucial step in confirming the success of the mutagenesis.
- Sequencing verification: Finally, sequencing the complete plasmid confirms the successful introduction of the intended mutation and rules out any unintended changes.
Troubleshooting Common Problems
Several issues can arise during SDM. Understanding these challenges and their potential solutions is crucial:
- No PCR product: This could be due to primer design flaws (incorrect Tm, secondary structures, or 3' mismatches), insufficient template DNA, or suboptimal PCR conditions.
- Non-specific amplification: This often stems from poor primer specificity (lack of BLAST analysis or high GC content).
- Low yield of mutated plasmid: This could result from inefficient primer annealing, inappropriate PCR conditions, or incomplete DpnI digestion.
Conclusion
Site-directed mutagenesis is a powerful technique for studying gene function, but its success hinges heavily on meticulous primer design. By carefully considering the factors outlined above, researchers can significantly improve the efficiency and reliability of SDM experiments. Utilizing online tools while critically evaluating the results ensures successful introduction of targeted mutations, leading to valuable insights in molecular biology research. Remember that while tools are helpful, a strong understanding of the underlying principles remains paramount for effective primer design and successful site-directed mutagenesis. Always perform careful analysis and verification steps to ensure accuracy and the validity of your results.
Latest Posts
Latest Posts
-
Is Water A Reactant Or Product
Mar 31, 2025
-
Four Ways To Represent A Function
Mar 31, 2025
-
What Is The Functional Unit Of Heredity
Mar 31, 2025
-
Cracking The Code Of Life Answer Key
Mar 31, 2025
-
Delta H Is Negative Exothermic Or Endothermic
Mar 31, 2025
Related Post
Thank you for visiting our website which covers about Primer Design For Site Directed Mutagenesis . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
