How Many Copies Of Dna Are Generated After 30 Cycles
Muz Play
Mar 20, 2025 · 5 min read
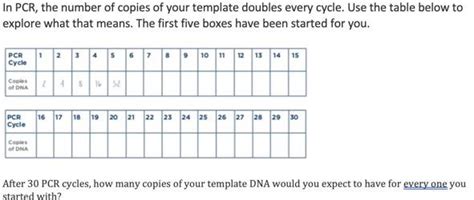
Table of Contents
How Many Copies of DNA are Generated After 30 Cycles of PCR? A Deep Dive into Exponential Amplification
Polymerase Chain Reaction (PCR) is a revolutionary technique that allows scientists to exponentially amplify a specific DNA sequence. Understanding the output of a PCR reaction, specifically the number of DNA copies generated after a given number of cycles, is crucial for various applications, from diagnostics to research. This article will delve into the mathematics behind PCR amplification, explore the factors that can influence the final copy number, and address common misconceptions surrounding this powerful technique.
The Power of Exponential Amplification: Understanding the 2<sup>n</sup> Rule
The core principle of PCR lies in its exponential amplification. Each cycle doubles the number of DNA copies. This means that if you start with one DNA molecule, after one cycle you'll have two, after two cycles you'll have four, and so on. This can be mathematically represented as 2<sup>n</sup>, where 'n' is the number of cycles.
Therefore, after 30 cycles of PCR, theoretically, you would have 2<sup>30</sup> copies of the target DNA sequence. This translates to a staggering 1,073,741,824 copies. This incredible amplification capacity is what makes PCR such a valuable tool.
The Ideal Scenario: Perfect Doubling in Every Cycle
The 2<sup>n</sup> formula represents the ideal scenario. In reality, PCR efficiency rarely reaches 100%. Various factors can influence the number of copies generated, leading to a deviation from this theoretical maximum.
Factors Affecting PCR Efficiency and Final Copy Number
Several factors can impact the actual number of DNA copies produced after 30 cycles:
1. Primer Efficiency and Specificity:
- Primer Design: Poorly designed primers can lead to low amplification efficiency. Factors like primer length, melting temperature (Tm), GC content, and potential self-complementarity significantly impact their ability to bind to the target DNA sequence. Inefficient primers will result in fewer copies being generated.
- Primer Dimers: Primers can sometimes anneal to each other instead of the target DNA, forming primer dimers. This reduces the available primers for amplification, leading to lower copy numbers.
- Specificity: Non-specific binding of primers to unintended sequences can lead to the amplification of unwanted products, consuming reagents and thus reducing amplification of the target DNA.
2. Enzyme Activity and Concentration:
- DNA Polymerase: The DNA polymerase enzyme is responsible for synthesizing new DNA strands. The quality and quantity of the polymerase directly affect the efficiency of the PCR reaction. A suboptimal enzyme or insufficient concentration can limit the amplification process.
- Mg<sup>2+</sup> Concentration: Magnesium ions are essential cofactors for DNA polymerase activity. An incorrect concentration of Mg<sup>2+</sup> can significantly impact enzyme function and, consequently, the number of copies generated.
3. Template DNA Quality and Concentration:
- DNA Integrity: Degraded or damaged template DNA will reduce the efficiency of PCR. The presence of inhibitors in the template DNA preparation can also negatively affect amplification.
- Template Concentration: Starting with too little template DNA can lead to insufficient amplification, while excessively high concentrations can lead to PCR inhibition.
4. Reaction Conditions and Optimization:
- Annealing Temperature: The annealing temperature is crucial for primer binding. An incorrect annealing temperature can lead to either low specificity (too low) or poor primer binding (too high), reducing the number of amplified copies.
- Extension Time: The extension time should be sufficient for the polymerase to synthesize the entire target sequence. Insufficient extension time can result in incomplete amplification.
- Cycling Parameters: The number of cycles is a significant factor. While 30 cycles are common, increasing the cycle number can eventually lead to a plateau where amplification efficiency decreases due to reagent depletion or accumulation of by-products.
5. Reagent Quality:
Using high-quality reagents, including dNTPs (deoxynucleotide triphosphates) and buffers, is essential for optimal PCR performance. Contaminated reagents can introduce inhibitors and reduce amplification efficiency.
The Plateau Effect: When Amplification Levels Off
Even with perfectly optimized conditions, PCR amplification eventually reaches a plateau. This is because:
- Reagent Depletion: The reaction mixture contains a limited amount of reagents, including dNTPs, primers, and polymerase. As the reaction progresses, these reagents are consumed, eventually limiting further amplification.
- Inhibitor Accumulation: Byproducts of the PCR reaction can accumulate, inhibiting further DNA synthesis.
Practical Considerations and Experimental Validation
While the theoretical calculation of 2<sup>30</sup> provides a useful estimate, it's crucial to remember that this rarely reflects the actual yield in a real-world PCR experiment. To accurately determine the number of copies generated, several methods can be employed:
- Quantitative PCR (qPCR): qPCR allows for precise quantification of DNA copies by measuring the amplification in real-time. This technique provides a much more accurate representation of the final copy number.
- Gel Electrophoresis: While not quantitative, gel electrophoresis can provide a visual assessment of the amplification product. The intensity of the band corresponding to the amplified DNA can be used to roughly compare amplification yields across different reactions.
- Spectrophotometry: Measuring the absorbance of DNA at 260nm can provide an estimate of the total DNA concentration, allowing for a calculation of the approximate copy number. However, this method requires careful consideration of purity and potential contamination.
Beyond the Numbers: Applications of PCR Amplification
The vast number of DNA copies generated through PCR has revolutionized various fields, including:
- Diagnostic testing: PCR is widely used for the detection and diagnosis of infectious diseases like COVID-19, influenza, and HIV.
- Forensic science: PCR enables the amplification of minute amounts of DNA from crime scenes, facilitating DNA profiling.
- Genetic research: PCR is indispensable in gene cloning, mutagenesis, and gene expression studies.
- Medicine: PCR is used in various medical applications, including prenatal diagnosis, cancer detection, and personalized medicine.
Conclusion: Understanding the Nuances of PCR Amplification
While the theoretical number of DNA copies generated after 30 PCR cycles is 2<sup>30</sup> (1,073,741,824), the actual yield is influenced by numerous factors. Achieving optimal PCR efficiency requires careful attention to primer design, enzyme selection, reaction conditions, and reagent quality. Accurate quantification of the final copy number often necessitates techniques like qPCR. Understanding these nuances is vital for reliable and meaningful results in diverse applications across scientific disciplines. The exponential amplification power of PCR remains a cornerstone of modern molecular biology, driving advancements in diagnostics, research, and countless other fields.
Latest Posts
Latest Posts
-
What Element Has 8 Protons And 8 Neutrons
Mar 20, 2025
-
The Law Of Sines Ambiguous Case
Mar 20, 2025
-
How To Find The Instantaneous Acceleration
Mar 20, 2025
-
Periodic Table Gases Liquids And Solids
Mar 20, 2025
-
The Dorsal Body Cavity Is Subdivided Into The
Mar 20, 2025
Related Post
Thank you for visiting our website which covers about How Many Copies Of Dna Are Generated After 30 Cycles . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
