Dichotomous Key For Gram Negative Bacteria
Muz Play
Mar 22, 2025 · 6 min read
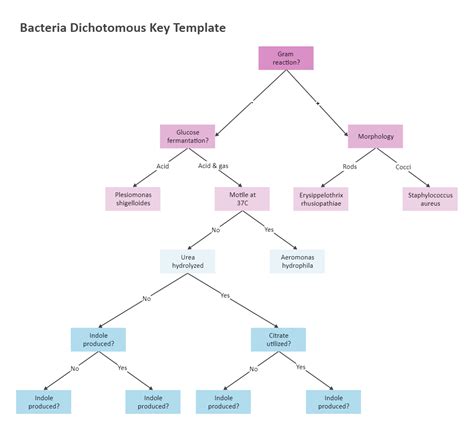
Table of Contents
Dichotomous Key for Gram-Negative Bacteria: A Comprehensive Guide
Gram-negative bacteria represent a vast and diverse group of microorganisms, encompassing both beneficial and pathogenic species. Accurate identification is crucial in various fields, including clinical diagnostics, environmental microbiology, and industrial applications. A dichotomous key serves as an invaluable tool for systematically identifying these bacteria, guiding users through a series of choices based on observable characteristics to ultimately pinpoint the genus and species. This comprehensive guide explores the principles behind constructing and utilizing a dichotomous key for gram-negative bacteria, highlighting key morphological, physiological, and biochemical features used for differentiation.
Understanding Dichotomous Keys
A dichotomous key, also known as a determinative key, is a hierarchical identification tool based on a series of paired contrasting statements, or couplets. Each couplet presents two mutually exclusive options, guiding the user down a specific path based on the characteristics of the unknown bacterium. The process continues until the organism is identified to a specific taxonomic level, usually genus and species.
Key features of a dichotomous key:
- Hierarchical structure: The key is organized in a hierarchical manner, branching out from broader characteristics to more specific traits.
- Paired statements (couplets): Each step presents two contrasting statements, requiring a decision based on observable characteristics.
- Sequential identification: The user progresses sequentially through the key, following the path determined by their observations.
- Terminal identification: The process concludes with the identification of the bacterium to a specific taxonomic level.
Essential Characteristics for Gram-Negative Bacteria Identification
Identifying gram-negative bacteria requires careful observation and testing of various characteristics. These characteristics fall into several broad categories:
1. Morphological Characteristics:
- Cell shape: Gram-negative bacteria exhibit various shapes, including cocci (spherical), bacilli (rod-shaped), vibrios (comma-shaped), spirilla (spiral-shaped), and spirochetes (flexible spiral-shaped). Microscopic examination is essential to determine the cell shape.
- Cell arrangement: Bacteria can occur singly, in pairs (diplococci, diplobacilli), chains (streptococci, streptobacilli), clusters (staphylococci), or other arrangements. This characteristic provides valuable clues for identification.
- Size and dimensions: Accurate measurement of bacterial size is important for differentiation, often requiring microscopic observation with a calibrated eyepiece.
- Presence of flagella: Flagella are whip-like appendages used for motility. The number, location (monotrichous, amphitrichous, lophotrichous, peritrichous), and arrangement of flagella are significant identification features.
- Presence of capsules: Some gram-negative bacteria possess capsules, which are gelatinous layers surrounding the cell wall. Capsule presence can be detected using specific staining techniques.
- Presence of endospores: Endospores are resistant structures formed by some bacteria under harsh conditions. While less common in gram-negative bacteria than in gram-positive, their presence is a significant identifying factor.
2. Physiological and Biochemical Characteristics:
- Oxygen requirements: Bacteria are classified based on their oxygen requirements as aerobes (require oxygen), anaerobes (cannot tolerate oxygen), facultative anaerobes (can grow with or without oxygen), and microaerophiles (require low oxygen concentrations). This characteristic can be assessed using different culture techniques.
- Nutrient requirements: Different bacteria have different nutritional needs. Growth on specific media (e.g., MacConkey agar, EMB agar) can be indicative of specific metabolic pathways and can be used in the key.
- Metabolic pathways: Various biochemical tests assess different metabolic pathways, including fermentation of sugars (glucose, lactose, sucrose), production of hydrogen sulfide (H2S), indole production, oxidase activity, catalase activity, and urease activity. These tests often form the backbone of a dichotomous key.
- Antibiotic susceptibility: The susceptibility of a bacterium to different antibiotics can be a valuable tool for identification. This often relies on antibiotic sensitivity testing, such as Kirby-Bauer disk diffusion.
- Temperature and pH optima: Bacteria exhibit optimal growth within specific temperature and pH ranges. These characteristics can be assessed through growth experiments under varying conditions.
- Motility: As mentioned above, motility is a key characteristic determined by the presence or absence of flagella. Motility can be visually observed using hanging drop preparations or motility agar.
3. Molecular Characteristics (Advanced Techniques):
While traditional methods are sufficient for many identifications, molecular techniques offer more precise and rapid identification. These advanced techniques include:
- 16S rRNA gene sequencing: This technique sequences a highly conserved gene (16S rRNA) present in all bacteria. Comparison with known sequences in databases allows for precise identification.
- DNA-DNA hybridization: This technique assesses the similarity of DNA sequences between two bacterial strains. High similarity suggests close taxonomic relatedness.
- Whole-genome sequencing: This advanced technique sequences the entire bacterial genome, providing extremely detailed information about the organism.
Constructing a Dichotomous Key for Gram-Negative Bacteria
Constructing a robust dichotomous key requires careful consideration of the relevant characteristics and their variability across different species. Here's a step-by-step approach:
-
Select representative species: Choose a representative selection of gram-negative bacteria you wish to identify. The selection should cover a range of morphological, physiological, and biochemical characteristics.
-
Identify key characteristics: Determine the most reliable and discriminating characteristics for differentiating the selected species. Begin with broad characteristics (e.g., cell shape) and then progress to more specific features (e.g., biochemical test results).
-
Develop couplets: Formulate pairs of contrasting statements (couplets) based on the chosen characteristics. Each couplet should lead to a subsequent step in the key or to the identification of a specific bacterium.
-
Organize the key: Arrange the couplets in a logical, hierarchical sequence. Begin with easily observable characteristics, leading progressively to more specific traits.
-
Test and refine: Thoroughly test the key using known bacterial cultures. If ambiguities or inaccuracies are identified, revise the key until it consistently provides accurate identifications.
Example of a Simplified Dichotomous Key
This is a greatly simplified example and would not be comprehensive enough for real-world applications. A true dichotomous key for gram-negative bacteria would be considerably more extensive.
1. Cell shape: a. Cocci – go to 2 b. Bacilli – go to 3
2. Arrangement of cocci: a. Diplococci – Neisseria b. Clusters – Staphylococcus (note: Staphylococcus is gram-positive, this is for illustrative purposes only)
3. Motility: a. Motile – go to 4 b. Non-motile – Escherichia coli (Note: E.coli is motile, this is for illustrative purposes only)
4. Lactose fermentation: a. Ferments lactose – Klebsiella (Note: Many lactose fermenters exist) b. Does not ferment lactose – Salmonella (Note: many non-lactose fermenters exist)
Conclusion
Dichotomous keys are invaluable tools for the identification of gram-negative bacteria. The ability to systematically differentiate between various species is crucial in many areas, including clinical diagnostics, environmental microbiology, and research. While the creation of a comprehensive dichotomous key for gram-negative bacteria requires extensive knowledge and expertise, the principles outlined in this guide provide a foundational understanding of this powerful identification technique. Remember that modern molecular techniques are increasingly being integrated into bacterial identification workflows, augmenting the capabilities of traditional methods. Combining classical approaches with modern molecular techniques ensures accurate and timely identification, leading to better understanding and management of these diverse and ecologically significant organisms.
Latest Posts
Latest Posts
-
Compared With Solid Ionic Compounds Solid Molecular Compounds Generally
Mar 22, 2025
-
Water Is Pure Substance Or Mixture
Mar 22, 2025
-
What Is A Complex Fraction In Math
Mar 22, 2025
-
Ba Oh 2 Strong Or Weak
Mar 22, 2025
-
Do Noble Gases Have Ionization Energy
Mar 22, 2025
Related Post
Thank you for visiting our website which covers about Dichotomous Key For Gram Negative Bacteria . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
