Gene Regulation In Eukaryotes Vs Prokaryotes
Muz Play
Mar 17, 2025 · 6 min read
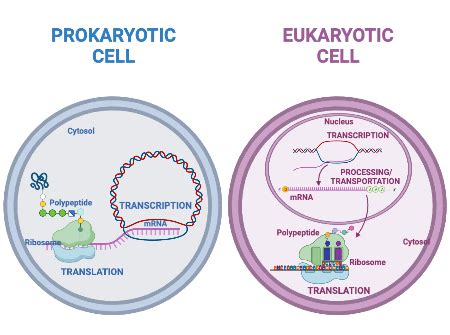
Table of Contents
Gene Regulation: A Tale of Two Domains – Eukaryotes vs. Prokaryotes
Gene regulation, the intricate orchestration of gene expression, is fundamental to life. However, the mechanisms governing this process differ significantly between prokaryotes (bacteria and archaea) and eukaryotes (plants, animals, fungi, and protists). Understanding these differences is key to comprehending the complexities of cellular function and evolution. This article delves deep into the fascinating world of gene regulation, comparing and contrasting the strategies employed by these two distinct domains of life.
The Prokaryotic Paradigm: Simple, Efficient, and Rapid Response
Prokaryotic gene regulation is characterized by its simplicity and speed. Prokaryotes, lacking the compartmentalization of eukaryotes, often regulate genes in coordinated groups called operons. This strategy allows for a rapid response to environmental changes. The most well-studied example is the lac operon in E. coli.
The lac Operon: A Classic Example
The lac operon controls the expression of genes involved in lactose metabolism. When lactose is absent, a repressor protein binds to the operator region, preventing RNA polymerase from transcribing the genes. However, when lactose is present, it binds to the repressor, causing a conformational change that prevents it from binding to the operator. This allows RNA polymerase to transcribe the genes, leading to lactose metabolism.
Other Regulatory Mechanisms in Prokaryotes
Beyond operons, prokaryotes utilize several other regulatory mechanisms:
-
Attenuation: This mechanism involves premature termination of transcription based on the availability of specific metabolites. For example, in the trp operon, tryptophan levels influence the formation of a secondary structure in the mRNA, leading to either transcription termination or continuation.
-
Global Regulators: These proteins, such as cAMP receptor protein (CRP), regulate the expression of multiple operons simultaneously, allowing for a coordinated response to environmental signals. CRP, for instance, is involved in regulating genes involved in carbon metabolism.
-
Two-Component Regulatory Systems: These systems involve a sensor kinase that detects environmental signals and a response regulator that modulates gene expression accordingly. These systems are commonly involved in responses to stress and changes in nutrient availability.
-
Riboswitches: These are structured RNA elements located in the 5' untranslated region of mRNA that bind to small molecules, directly affecting translation initiation. This allows for direct regulation based on the presence or absence of specific metabolites.
In essence, prokaryotic gene regulation is characterized by its responsiveness to environmental cues, its reliance on operons for coordinated gene expression, and the direct coupling of transcription and translation. The lack of a nucleus and the presence of a single circular chromosome contribute to the streamlined nature of this regulation.
The Eukaryotic Enigma: Complex, Multilayered, and Precise Control
Eukaryotic gene regulation is significantly more complex than its prokaryotic counterpart. This complexity arises from several factors, including:
- Compartmentalization: The presence of a nucleus separates transcription from translation, allowing for multiple levels of control.
- Chromatin Structure: DNA is packaged into chromatin, influencing the accessibility of genes to transcriptional machinery.
- Multiple RNA Processing Steps: Eukaryotic mRNA undergoes several processing steps, including capping, splicing, and polyadenylation, providing additional layers of regulation.
Levels of Eukaryotic Gene Regulation
Eukaryotic gene regulation occurs at multiple levels:
-
Chromatin Remodeling: This involves changes in chromatin structure that alter the accessibility of DNA to transcription factors. Histone modifications (e.g., acetylation, methylation) and chromatin remodeling complexes play crucial roles in this process.
-
Transcriptional Regulation: This is the most extensively studied level of regulation and involves the binding of transcription factors to specific DNA sequences (promoters and enhancers) to either activate or repress transcription. Transcription factors are proteins that interact with RNA polymerase and other regulatory elements to control the rate of transcription.
-
RNA Processing: This includes the addition of a 5' cap, splicing of introns, and the addition of a poly(A) tail. These processes can significantly impact mRNA stability and translation efficiency. Alternative splicing, where different combinations of exons are joined, can lead to the production of multiple protein isoforms from a single gene.
-
mRNA Transport and Localization: The transport of mRNA from the nucleus to the cytoplasm and its localization within the cell can also be regulated.
-
Translational Regulation: This involves controlling the rate at which mRNA is translated into protein. Factors affecting translation initiation, elongation, and termination can modulate protein synthesis.
-
Post-Translational Modification: This includes various processes like phosphorylation, glycosylation, and ubiquitination, which can alter protein activity, stability, and localization.
Key Players in Eukaryotic Gene Regulation
Several key players contribute to the intricate dance of eukaryotic gene regulation:
- Transcription Factors: These proteins bind to specific DNA sequences and interact with RNA polymerase to either activate or repress transcription.
- Histone Modifying Enzymes: These enzymes add or remove chemical groups from histones, influencing chromatin structure and gene accessibility.
- Chromatin Remodeling Complexes: These multiprotein complexes alter nucleosome position and spacing, influencing DNA accessibility.
- RNA Binding Proteins: These proteins bind to RNA molecules and influence RNA processing, stability, and translation.
- miRNAs and siRNAs: These small non-coding RNAs regulate gene expression by binding to target mRNAs, leading to mRNA degradation or translational repression.
Comparing and Contrasting: A Summary Table
| Feature | Prokaryotes | Eukaryotes |
|---|---|---|
| Transcription/Translation Coupling | Coupled | Uncoupled |
| Genome Structure | Single, circular chromosome | Multiple, linear chromosomes |
| Gene Organization | Operons common | Operons rare |
| Chromatin Structure | No nucleosomes | Nucleosomes present |
| Transcription Regulation | Primarily at the initiation level | Multiple levels (chromatin, transcription, RNA processing, translation, post-translation) |
| RNA Processing | Minimal processing | Extensive processing (capping, splicing, polyadenylation) |
| Response Time | Rapid | Slower |
| Complexity | Relatively simple | Highly complex |
| Regulatory Mechanisms | Operons, attenuation, global regulators, two-component systems, riboswitches | Chromatin remodeling, transcription factors, RNA processing, RNA interference, translational control, post-translational modifications |
The Evolutionary Perspective: Adaptation and Complexity
The differences in gene regulation between prokaryotes and eukaryotes reflect their distinct evolutionary trajectories. Prokaryotes, often facing rapidly changing environments, require quick and efficient responses to environmental stimuli. Their simple regulatory mechanisms, such as operons and two-component systems, provide this necessary speed.
Eukaryotes, with their greater cellular complexity and longer lifespans, require more precise and nuanced control over gene expression. The multilayered regulatory mechanisms employed by eukaryotes, including chromatin remodeling, RNA processing, and post-translational modifications, allow for greater control over gene expression, enabling complex developmental processes and tissue differentiation. The evolution of the nucleus and the separation of transcription and translation likely played a crucial role in the development of this complexity.
Future Directions: Unraveling the Intricacies
Despite significant advances in our understanding of gene regulation, much remains to be discovered. Ongoing research focuses on:
- Uncovering novel regulatory mechanisms: Advanced technologies, such as high-throughput sequencing and CRISPR-Cas9 gene editing, are revealing new players and interactions in gene regulatory networks.
- Understanding the interplay between different regulatory levels: Researchers are investigating the intricate crosstalk between different levels of regulation, such as the coordination of chromatin remodeling and transcription.
- Developing therapeutic applications: A deeper understanding of gene regulation holds enormous potential for the development of new therapies for diseases such as cancer and genetic disorders. Targeted manipulation of gene expression offers a promising avenue for future treatments.
The comparison of gene regulation in prokaryotes and eukaryotes reveals a fascinating evolutionary story. The simple, rapid responses of prokaryotes contrast sharply with the intricate, multifaceted control mechanisms of eukaryotes. Continued research promises to further illuminate the complexity and elegance of gene regulation in both domains of life, ultimately advancing our understanding of fundamental biological processes and opening new avenues for therapeutic intervention.
Latest Posts
Latest Posts
-
The Ideal Osmotic Environment For An Animal Cell Is
Mar 17, 2025
-
What Are The Three Basic Components Of An Atom
Mar 17, 2025
-
Does Gas Have A Definite Shape
Mar 17, 2025
-
If The Equilibrium Constant Is Negative What Does That Mean
Mar 17, 2025
-
How Does An Atom Become A Cation
Mar 17, 2025
Related Post
Thank you for visiting our website which covers about Gene Regulation In Eukaryotes Vs Prokaryotes . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
